|
Evaluating the performance of methods for inference of gene regulatory networks is difficult, because network predictions can in general not be systematically tested in vivo with the current technology. Consequently, in silico benchmarks based on simulated networks and expression data are essential to assess the performance of inference methods (Figure 1).
We are developing tools for the generation of biologically plausible benchmarks, which enable realistic in silico performance assessment:
We generate realistic network structures for the benchmarks by extracting modules from known gene networks of model organisms, instead of using random graph models as commonly done. [2] We endow these networks with dynamics using a kinetic model that is more detailed than those of other in silico benchmarks. It is based on a thermodynamical model of transcriptional regulation, and it includes both mRNA and protein dynamics. Furthermore, in addition to measurement noise, we also model internal noise in the dynamics of the gene networks. [1]
This framework allows inference methods to be tested in silico on networks with similar types of structural properties, regulatory dynamics, and noise as occur in biological gene networks.
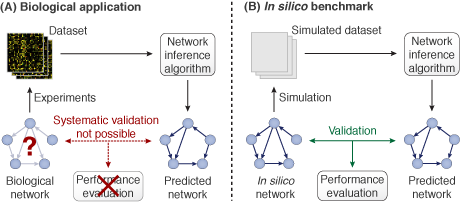
Figure 1. (A) The "true" network structure of biological gene networks is in general unknown, which hinders systematic performance evaluation. (B) For in silico networks, the ground truth is known and predictions can be validated.
|